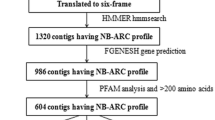
Abstract
NBS-encoding genes play a critical role in the plant defense system. Wild relatives of crop plants are rich reservoirs of plant defense genes. Here, we performed a stringent genome-wide identification of NBS-encoding genes in three cultivated and eight wild Oryza species, representing three different genomes (AA, BB, and FF) from four continents. A total of 2688 NBS-encoding genes were identified from 11 Oryza genomes. All the three progenitor species of cultivated rice, namely O. barthii, O. rufipogon, and O. nivara, were the richest reservoir of NBS-encoding genes (214, 313, and 307 respectively). Interestingly, the two Asian cultivated species showed a contrasting pattern in the number of NBS-encoding genes. While indica subspecies maintained nearly equal number of NBS genes as its progenitor (309 and 313), the japonica subspecies had retained only two third in the course of evolution (213 and 307). Other major sources for NBS-encoding genes could be (i) O. longistaminata since it had the highest proportion of NBS-encoding genes and (ii) O. glumaepatula as it clustered distinctly away from the rest of the AA genome species. The present study thus revealed that NBS-encoding genes can be exploited from the primary gene pool for disease resistance breeding in rice.







Similar content being viewed by others
References
Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Ameline-Torregrosa C, Wang BB, O’Bleness MS et al (2008) Identification and characterization of NBS–LRR genes in the model plant Medicago truncatula. Plant Physiol 146:5–21
Andolfo G, Jupe F, Witek K, Etherington GJ, Ercolano MR, Jones JD (2014) Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol 14:120. https://doi.org/10.1186/1471-2229-14-120
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinf 10:421
Cheng X, Jiang H, Zhao Y, Qian Y, Zhu S, Cheng B (2009) A genomic analysis of disease-resistance genes encoding nucleotide binding sites in Sorghum bicolor. Genet Mol Biol 33(2):292–297. https://doi.org/10.1590/S1415-47572010005000036
Das A, Soubam D, Singh PK, Thakur S, Singh NK, Sharma TR (2012) A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct Integr Genomics 12:215–228. https://doi.org/10.1007/s10142-012-0284-1
Devanna NB, Vijayan J, Sharma TR (2014) The Blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS One 9(8):e104840. https://doi.org/10.1371/journal.pone.0104840
Guo YL, Fitz J, Schneeberger K, Ossowski S, Cao J, Weigel D (2011) Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol 157:757–769. https://doi.org/10.1104/pp.111.181990
He Z, Zhai W, Wen H, Tang T, Wang Y, Lu X, Greenberg AJ, Hudson RR, Wu CI, Shi S (2011) Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet 7:e1002100. https://doi.org/10.1371/journal.pgen.1002100
Heidel AJ, Lawal HM, Felder M et al (2011) Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res 21:1882–1891. https://doi.org/10.1101/gr.121137.111
Holub EB (2001) The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2:516–527
Hulsen T, Huynen MA, J-de V, Groenen PMA (2006) Benchmarking ortholog identification methods using functional genomics data. Genome Biol 7:R31. https://doi.org/10.1186/gb-2006-7-4-r31
Jaillon O, Aury JM, Noel B et al (2001) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Joshi RK, Nayak S (2013) Perspectives of genomic diversification and molecular recombination towards R-gene evolution in plants. Physiol Mol Biol Plants 19:1–9
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:24–34
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404.
Li J, Ding J, Zhang W, Zhang Y, Tang P, Chen JQ, Tian D, Yang S (2010) Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol Gen Genomics 283:427–438. https://doi.org/10.1007/s00438-010-0527-6
Liu W, Liu J, Triplett L, Leach JE, Wang GL (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 52:213–241. https://doi.org/10.1146/annurev-phyto-102313-045926
Lozano R, Hamblin MT, Prochnik S, Jannink JL (2015) Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics 16:360. https://doi.org/10.1186/s12864-015-1554-9
Lukasik E, Takken FL (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12:427–436. https://doi.org/10.1016/j.pbi.2009.03.001
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39(Database issue):D225–D229
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834. https://doi.org/10.1105/tpc.009308
Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD, Boyko AR, Purugganan MD (2011) Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci 108:8351–8356
Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, Meyers BC (2013) The role of TIR-NBS and TIR-X proteins in plant basal defence responses. Plant Physiol 162:1459–1472. https://doi.org/10.1104/pp.113.219162
Nguyen BD, Brar DS, Bui BC, Nguyen TV, Pham LN, Nguyen HT (2013) Identification and mapping of the QTL for aluminum tolerance introgressed from new source, Oryza rufipogon Griff. into indica rice, (Oryza sativa L.). Theor Appl Genet 106:583–593
Park KC, Kim NH, Cho YS, Kang KH, Lee JK, Kim NS (2003) Genetic variations of AA genome Oryza species measured by MITE-AFLP. Theor Appl Genet 107:203–209
Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Gen Genomics 281:609–626. https://doi.org/10.1007/s00438-009-0434-x
Rahman ML, Jiang W, Chu SH, Qiao Y, Ham TH, Woo MO, Lee J, Khanam MS, Chin JH, Jeung JU, Brar DS, Jena KK, Koh HJ (2009) High-resolution mapping of two rice brown plant hopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor Appl Genet 119:1237–1246
Ram T, Majumder ND, Krishnaveni D, Ansari MM (2007) Rice variety Dhanarasi, an example of improving yield potential and disease resistance by introgressing gene(s) from wild species (O. rufipogon). Curr Sci 92:987–992
Rawal HC, Singh NK, Sharma TR (2013) Conservation, divergence and genome wide distribution of PAL and POX A gene families in plants. Int J Genomics 13:1–10. https://doi.org/10.1155/2013/678969
Sanchez PL, Wing RA, Brar DS. 2014 The wild relative of Rice: genomes and genomics, in Genetics and genomics of rice, ed. Zhang Q. and Wing R.A. (Plant genetics and genomics: crops and models 5, Springer), 9–25. DOI https://doi.org/10.1007/978-1-4614-7903-1_2
Second G (1982) Origin of the genetic diversity of cultivated rice (Oryza spp.): study of the polymorphism scored at 40 isozyme loci. Jpn J Genet 57:25–57
Seo E, Kim S, Yeom SI, Choi D (2016) Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among Solanaceae plants. Front Plant Sci 7:1205. https://doi.org/10.3389/fpls.2016.01205
Sharma T, Rai A, Gupta S, Vijayan J, Devanna B, Ray S (2012) Rice blast management through host-plant resistance: retrospect and prospects. Agric Res 1(1):37–52. https://doi.org/10.1007/s40003-011-0003-5
Singh S, Chand S, Singh NK, Sharma TR (2015) Genome-wide distribution, organisation and functional characterization of disease resistance and defence response genes across rice species. PLoS One 10(4):e0125964. https://doi.org/10.1371/journal.pone.0125964
Tanweer FA, Rafii MY, Sijam K, Rahim HA, Ahmed F, Latif MA (2015) Current advance methods for the identification of blast resistance genes in rice. C R Biol 338:321–334. https://doi.org/10.1016/j.crvi.2015.03.001
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, dePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, van de Peer Y, Rokhsar D (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Van der Biezen EA, Jones JD (1998) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8(7):R226–R227
Van Zee JP, Schlueter JA, Schlueter S, Dixon P, Sierra CA, Hill CA (2016) Paralog analyses reveal gene duplication events and genes under positive selection in Ixodes scapularis and other ixodid ticks. BMC Genomics 17:241. https://doi.org/10.1186/s12864-015-2350-2
Vaughan DA (1994) The wild relatives of rice: a genetic resources handbook. International Rice Research Institute, Manila, p 137
Vaughan DA, Lu BR, Tomooka N (2008) The evolving story of rice evolution. Plant Sci 174:394–408
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ (2015) Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci Rep 5:13957. https://doi.org/10.1038/srep13957
Waters DL, Nock CJ, Ishikawa R, Rice N, Henry RJ (2012) Chloroplast genome sequence confirms distinctness of Australian and Asian wild rice. Ecol Evol 2:211–217. https://doi.org/10.1002/ece3.66
Yang S, Zhang X, Yue JX, Tian D, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Gen Genomics 280:187–198. https://doi.org/10.1007/s00438-008-0355-0
Yue JX, Meyers BC, Chen JQ, Tian DC, Yang SH (2012) Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol 193:1049–1063. https://doi.org/10.1111/j.1469-8137.2011.04006.x
Zhang QJ, Zhu T, Xia EH, Shi C, Liu YL, Zhang Y, Liu Y, Jiang WK, Zhao YJ, Mao SY, Zhang LP, Huang H, Jiao JY, Xu PZ, Yao QY, Zeng FC, Yang LL, Gao J, Tao DY, Wang YJ, Bennetzen JL, Gao LZ (2014) Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc Natl Acad Sci USA 111:E4954–E4962. https://doi.org/10.1073/pnas.1418307111
Funding
This research received financial support from the Centre for Agricultural Bioinformatics Scheme (CABin) funded by the Indian Council of Agricultural Research (ICAR), New Delhi, India.
Author information
Authors and Affiliations
Contributions
AS and AM: conceived and designed the experiments; HR and NG: developed the pipelines and analyzed data; KA and VK: PCR validation; HR and AS: finalized tables and figures; DM, KC, and AR: provided the bioinformatic support; VD: supplied plant material; AM, AS, and TS: drafted and finalized the manuscript; and all the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Rawal, H.C., Amitha Mithra, S.V., Arora, K. et al. Genome-Wide Analysis in Wild and Cultivated Oryza Species Reveals Abundance of NBS Genes in Progenitors of Cultivated Rice. Plant Mol Biol Rep 36, 373–386 (2018). https://doi.org/10.1007/s11105-018-1086-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-018-1086-y